Centromere

The centromere is the specialized DNA sequence of a chromosome that links a pair of sister chromatids (a dyad).[1] During mitosis, spindle fibers attach to the centromere via the kinetochore.[2] Centromeres were first thought to be genetic loci that direct the behavior of chromosomes.
The physical role of the centromere is to act as the site of assembly of the kinetochores – a highly complex multiprotein structure that is responsible for the actual events of chromosome segregation – i.e. binding microtubules and signalling to the cell cycle machinery when all chromosomes have adopted correct attachments to the spindle, so that it is safe for cell division to proceed to completion and for cells to enter anaphase.[3]
There are, broadly speaking, two types of centromeres. "Point centromeres" bind to specific proteins that recognize particular DNA sequences with high efficiency.[4] Any piece of DNA with the point centromere DNA sequence on it will typically form a centromere if present in the appropriate species. The best characterised point centromeres are those of the budding yeast, Saccharomyces cerevisiae. "Regional centromeres" is the term coined to describe most centromeres, which typically form on regions of preferred DNA sequence, but which can form on other DNA sequences as well.[4] The signal for formation of a regional centromere appears to be epigenetic. Most organisms, ranging from the fission yeast Schizosaccharomyces pombe to humans, have regional centromeres.
Regarding mitotic chromosome structure, centromeres represent a constricted region of the chromosome (often referred to as the primary constriction) where two identical sister chromatids are most closely in contact. When cells enter mitosis, the sister chromatids (the two copies of each chromosomal DNA molecule resulting from DNA replication in chromatin form) are linked along their length by the action of the cohesin complex. It is now believed that this complex is mostly released from chromosome arms during prophase, so that by the time the chromosomes line up at the mid-plane of the mitotic spindle (also known as the metaphase plate), the last place where they are linked with one another is in the chromatin in and around the centromere.[5]
Position[edit]
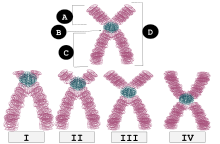
| I | Telocentric | Centromere placement very close to the top, p arms barely visible if visible at all. |
| II | Acrocentric | q arms are still much longer than the p arms, but the p arms are longer than those in telocentric. |
| III | Submetacentric | p and q arms are very close in length but not equal. |
| IV | Metacentric | p and q arms are equal in length. |
B: Centromere
C: Long arm (q arm)
D: Sister Chromatids
Each chromosome has two arms, labeled p (the shorter of the two) and q (the longer). Many remember that the short arm 'p' is named for the French word "petit" meaning 'small', although this explanation was shown to be apocryphal.[6] They can be connected in either metacentric, submetacentric, acrocentric or telocentric manner.[7][8]
| Categorization of chromosomes according to the relative arms length[8] | ||||||
| Centromere position | Arms length ratio | Sign | Description | |||
| Medial sensu stricto | 1.0 – 1.6 | M | Metacentric | |||
| Medial region | 1.7 | m | Metacentric | |||
| Submedial | 3.0 | sm | Submetacentric | |||
| Subterminal | 3.1 – 6.9 | st | Subtelocentric | |||
| Terminal region | 7.0 | t | Acrocentric | |||
| Terminal sensu stricto | ∞ | T | Telocentric | |||
| Notes | – | Metacentric: M+m | Atelocentric: M+m+sm+st+t | |||
Metacentric[edit]
These are X-shaped chromosomes, with the centromere in the middle so that the two arms of the chromosomes are almost equal.
A chromosome is metacentric if its two arms are roughly equal in length. In a normal human karyotype, five chromosomes are considered metacentric: chromosomes 1, 3, 16, 19, and 20. In some cases, a metacentric chromosome is formed by balanced translocation: the fusion of two acrocentric chromosomes to form one metacentric chromosome.[9][10]
Submetacentric[edit]
If the arms' lengths are unequal, the chromosome is said to be submetacentric. They are L-shaped.[11]
Acrocentric[edit]
If the p (short) arm is so short that it is hard to observe, but still present, then the chromosome is acrocentric (the "acro-" in acrocentric refers to the Greek word for "peak"). The human genome includes five acrocentric chromosomes: 13, 14, 15, 21, 22.[12] The Y chromosome is also acrocentric.[12]
In an acrocentric chromosome the p arm contains genetic material including repeated sequences such as nucleolar organizing regions, and can be translocated without significant harm, as in a balanced Robertsonian translocation. The domestic horse genome includes one metacentric chromosome that is homologous to two acrocentric chromosomes in the conspecific but undomesticated Przewalski's horse.[13] This may reflect either fixation of a balanced Robertsonian translocation in domestic horses or, conversely, fixation of the fission of one metacentric chromosome into two acrocentric chromosomes in Przewalski's horses. A similar situation exists between the human and great ape genomes, with a reduction of two acrocentric chromosomes in the great apes to one metacentric chromosome in humans (see aneuploidy and the human chromosome 2).[11]
Strikingly, harmful translocations in disease context, especially unbalanced translocations in blood cancers, more frequently involve acrocentric chromosomes than non-acrocentric chromosomes.[14] Although the cause is not known, this probably relates to the physical location of acrocentric chromosomes within the nucleus. Acrocentric chromosomes are usually located in and around the nucleolus, so in the center of the nucleus, where chromosomes tend to be less densely packed than chromosomes in the nuclear periphery.[15] Consistently, chromosomal regions that are less densely packed are also more prone to chromosomal translocations in cancers.[14]
Telocentric[edit]
A telocentric chromosome's centromere is located at the terminal end of the chromosome. A telocentric chromosome has therefore only one arm. Telomeres may extend from both ends of the chromosome, their shape is similar to letter "i" during anaphase. For example, the standard house mouse karyotype has only telocentric chromosomes.[16][17] Humans do not possess telocentric chromosomes.
Subtelocentric[edit]
If the chromosome's centromere is located closer to its end than to its center, it may be described as subtelocentric.[18][19]
Centromere number[edit]
Acentric[edit]
If a chromosome lacks a centromere, it is said acentric. The macronucleus of ciliates for example contains hundreds of acentric chromosomes.[20] Chromosome-breaking events can also generate acentric chromosomes or acentric fragments.
Dicentric[edit]
A dicentric chromosome is an abnormal chromosome with two centromeres. It is formed through the fusion of two chromosome segments, each with a centromere, resulting in the loss of acentric fragments (lacking a centromere) and the formation of dicentric fragments.[21] The formation of dicentric chromosomes has been attributed to genetic processes, such as Robertsonian translocation[12] and paracentric inversion.[22] Dicentric chromosomes have important roles in the mitotic stability of chromosomes and the formation of pseudodicentric chromosomes.[23]
Monocentric[edit]
The monocentric chromosome is a chromosome that has only one centromere in a chromosome and forms a narrow constriction.
Monocentric centromeres are the most common structure on highly repetitive DNA in plants and animals.[24]
Holocentric[edit]
Different than monocentric chromosomes in holocentric chromosomes, the entire length of the chromosome acts as the centromere. In holocentric chromosomes there is not one primary constriction but the centromere has many CenH3 loci spread over the whole chromosome.[25] Examples of this type of centromere can be found scattered throughout the plant and animal kingdoms,[26] with the most well-known example being the nematode Caenorhabditis elegans.
Polycentric[edit]
Human chromosomes[edit]
| Chromosome | Centromere position (Mbp) | Category | Chromosome Size (Mbp) | Centromere size (Mbp) |
|---|---|---|---|---|
| 1 | 125.0 | metacentric | 247.2 | 7.4 |
| 2 | 93.3 | submetacentric | 242.8 | 6.3 |
| 3 | 91.0 | metacentric | 199.4 | 6.0 |
| 4 | 50.4 | submetacentric | 191.3 | — |
| 5 | 48.4 | submetacentric | 180.8 | — |
| 6 | 61.0 | submetacentric | 170.9 | — |
| 7 | 59.9 | submetacentric | 158.8 | — |
| 8 | 45.6 | submetacentric | 146.3 | — |
| 9 | 49.0 | submetacentric | 140.4 | — |
| 10 | 40.2 | submetacentric | 135.4 | — |
| 11 | 53.7 | submetacentric | 134.5 | — |
| 12 | 35.8 | submetacentric | 132.3 | — |
| 13 | 17.9 | acrocentric | 114.1 | — |
| 14 | 17.6 | acrocentric | 106.3 | — |
| 15 | 19.0 | acrocentric | 100.3 | — |
| 16 | 36.6 | metacentric | 88.8 | — |
| 17 | 24.0 | submetacentric | 78.7 | — |
| 18 | 17.2 | submetacentric | 76.1 | — |
| 19 | 26.5 | metacentric | 63.8 | — |
| 20 | 27.5 | metacentric | 62.4 | — |
| 21 | 13.2 | acrocentric | 46.9 | — |
| 22 | 14.7 | acrocentric | 49.5 | — |
| X | 60.6 | submetacentric | 154.9 | — |
| Y | 12.5 | acrocentric | 57.7 | — |
Sequence[edit]
There are two types of centromeres.[27] In regional centromeres, DNA sequences contribute to but do not define function. Regional centromeres contain large amounts of DNA and are often packaged into heterochromatin. In most eukaryotes, the centromere's DNA sequence consists of large arrays of repetitive DNA (e.g. satellite DNA) where the sequence within individual repeat elements is similar but not identical. In humans, the primary centromeric repeat unit is called α-satellite (or alphoid), although a number of other sequence types are found in this region.[28]
Point centromeres are smaller and more compact. DNA sequences are both necessary and sufficient to specify centromere identity and function in organisms with point centromeres. In budding yeasts, the centromere region is relatively small (about 125 bp DNA) and contains two highly conserved DNA sequences that serve as binding sites for essential kinetochore proteins.[28]
Inheritance[edit]
Since centromeric DNA sequence is not the key determinant of centromeric identity in metazoans, it is thought that epigenetic inheritance plays a major role in specifying the centromere.[29] The daughter chromosomes will assemble centromeres in the same place as the parent chromosome, independent of sequence. It has been proposed that histone H3 variant CENP-A (Centromere Protein A) is the epigenetic mark of the centromere.[30] The question arises whether there must be still some original way in which the centromere is specified, even if it is subsequently propagated epigenetically. If the centromere is inherited epigenetically from one generation to the next, the problem is pushed back to the origin of the first metazoans.
Structure[edit]
The centromeric DNA is normally in a heterochromatin state, which is essential for the recruitment of the cohesin complex that mediates sister chromatid cohesion after DNA replication as well as coordinating sister chromatid separation during anaphase. In this chromatin, the normal histone H3 is replaced with a centromere-specific variant, CENP-A in humans.[31] The presence of CENP-A is believed to be important for the assembly of the kinetochore on the centromere. CENP-C has been shown to localise almost exclusively to these regions of CENP-A associated chromatin. In human cells, the histones are found to be most enriched for H4K20me3 and H3K9me3[32] which are known heterochromatic modifications. In Drosophila, Islands of retroelements are major components of the centromeres. [33]
In the yeast Schizosaccharomyces pombe (and probably in other eukaryotes), the formation of centromeric heterochromatin is connected to RNAi.[34] In nematodes such as Caenorhabditis elegans, some plants, and the insect orders Lepidoptera and Hemiptera, chromosomes are "holocentric", indicating that there is not a primary site of microtubule attachments or a primary constriction, and a "diffuse" kinetochore assembles along the entire length of the chromosome.
Centromeric aberrations[edit]
In rare cases in humans, neocentromeres can form at new sites on the chromosome. There are currently over 90 known human neocentromeres identified on 20 different chromosomes.[35][36] The formation of a neocentromere must be coupled with the inactivation of the previous centromere, since chromosomes with two functional centromeres (Dicentric chromosome) will result in chromosome breakage during mitosis. In some unusual cases human neocentromeres have been observed to form spontaneously on fragmented chromosomes. Some of these new positions were originally euchromatic and lack alpha satellite DNA altogether.
Centromere proteins are also the autoantigenic target for some anti-nuclear antibodies, such as anti-centromere antibodies.
Dysfunction and disease[edit]
It has been known that centromere misregulation contributes to mis-segregation of chromosomes, which is strongly related to cancer and abortion. Notably, overexpression of many centromere genes have been linked to cancer malignant phenotypes. Overexpression of these centromere genes can increase genomic instability in cancers.[37] Elevated genomic instability on one hand relates to malignant phenotypes; on the other hand, it makes the tumor cells more vulnerable to specific adjuvant therapies such as certain chemotherapies and radiotherapy.[38] Instability of centromere repetitive DNA was recently shown in cancer and aging.[39]
Etymology and pronunciation[edit]
The word centromere (/ˈsɛntrəˌmɪər/[40][41]) uses combining forms of centro- and -mere, yielding "central part", describing the centromere's location at the center of the chromosome.
See also[edit]
References[edit]
- ^ Alberts, Bruce; Bray, Dennis; Hopkin, Karen; Johnson, Alexander; Lewis, Julian; Raff, Martin; Roberts, Keith; Walter, Peter (2014). Essential Cell Biology (4 ed.). New York, NY: Garland Science. p. 183. ISBN 978-0-8153-4454-4.
- ^ Pollard, T.D. (2007). Cell Biology. Philadelphia: Saunders. pp. 200–203. ISBN 978-1-4160-2255-8.
- ^ Pollard, TD (2007). Cell Biology. Philadelphia: Saunders. pp. 227–230. ISBN 978-1-4160-2255-8.
- ^ a b Pluta, A.; A.M. Mackay; A.M. Ainsztein; I.G. Goldberg; W.C. Earnshaw (1995). "The centromere: Hub of chromosomal activities". Science. 270 (5242): 1591–1594. Bibcode:1995Sci...270.1591P. doi:10.1126/science.270.5242.1591. PMID 7502067.
- ^ "Sister chromatid cohesion". Genetics Home Reference. United States National Library of Medicine. May 15, 2011.
- ^ "p + q = Solved, Being the True Story of How the Chromosome Got Its Name". 2011-05-03.
- ^ Nikolay's Genetics Lessons (2013-10-12), What different types of chromosomes exist?, retrieved 2017-05-28
- ^ a b Levan A., Fredga K., Sandberg A. A. (1964): Nomenclature for centromeric position on chromosomes. Hereditas, Lund, 52: 201.
- ^ "Chromosomes, Chromosome Anomalies".
- ^ *Gilbert F (1999). "Disease genes and chromosomes: disease maps of the human genome. Chromosome 16". Genet Test. 3 (2): 243–54. doi:10.1089/gte.1999.3.243. PMID 10464676.
- ^ a b Nussbaum, Robert L.; McInnes, Roderick R.; Thompson, Margaret Wilson; Thompson, James Scott; Willard, Huntington F. (2001). Thompson & Thompson Genetics in Medicine. ISBN 0721669026.
- ^ a b c Thompson & Thompson GENETICS IN MEDICINE 7th Edition. p. 62.
- ^ Myka, J.L.; Lear, T.L.; Houck, M.L.; Ryder, O.A.; Bailey, E. (2003). "FISH analysis comparing genome organization in the domestic horse (Equus caballus) to that of the Mongolian wild horse (E. przewalskii)". Cytogenetic and Genome Research. 102 (1–4): 222–5. doi:10.1159/000075753. PMID 14970707.
- ^ a b Lin, C.Y.; Shukla, A.; Grady, J.P.; Fink, J.L.; Dray, E.; Duijf, P.H.G. (2018), "Translocation breakpoints preferentially occur in euchromatin and acrocentric chromosomes", Cancers (Basel), 10 (1): E13, doi:10.3390/cancers10010013, PMC 5789363, PMID 29316705
- ^ Bolzer, A.; et, al. (2005), "Three-dimensional maps of all chromosomes in human male fibroblast nuclei and prometaphase rosettes.", PLOS Biology, 3 (5): e157, doi:10.1371/journal.pbio.0030157, PMC 1084335, PMID 15839726
- ^ Silver, Lee M. (1995). "Karyotypes, Chromosomes, and Translocations". Mouse Genetics: Concepts and Applications. Oxford: Oxford University Press. pp. 83–92. ISBN 978-0-19-507554-0.
- ^ Chinwalla, Asif T.; Cook, Lisa L.; Delehaunty, Kimberly D.; Fewell, Ginger A.; Fulton, Lucinda A.; Fulton, Robert S.; Graves, Tina A.; Hillier, Ladeana W.; et al. (2002). "Initial sequencing and comparative analysis of the mouse genome". Nature. 420 (6915): 520–62. Bibcode:2002Natur.420..520W. doi:10.1038/nature01262. PMID 12466850.
- ^ "subtelocentric chromosome definition". groups.molbiosci.northwestern.edu. Retrieved 2017-10-29.
- ^ Margulis, Lynn; Matthews, Clifford; Haselton, Aaron (2000-01-01). Environmental Evolution: Effects of the Origin and Evolution of Life on Planet Earth. MIT Press. ISBN 9780262631976.
- ^ Pevsner, Jonathan (2015-08-17). Bioinformatics and Functional Genomics. John Wiley & Sons. ISBN 9781118581766.
- ^ Nussbaum, Robert; McInnes, Roderick; Willard, Huntington; Hamosh, Ada (2007). Thompson & Thompson Genetics in Medicine. Philadelphia(PA): Saunders. p. 72. ISBN 978-1-4160-3080-5.
- ^ Hartwell, Leland; Hood, Leeroy; Goldberg, Michael; Reynolds, Ann; Lee, Silver (2011). Genetics From Genes to Genomes, 4e. New York: McGraw-Hill. ISBN 9780073525266.
- ^ Lynch, Sally; et al. (1995). "Kabuki syndrome-like features in monozygotic twin boys with a pseudodicentric chromosome 13". J. Med. Genet. 32 (32:227–230): 227–230. doi:10.1136/jmg.32.3.227. PMC 1050324. PMID 7783176.
- ^ Barra, V.; Fachinetti, D. (2018). "The dark side of centromeres: Types, causes and consequences of structural abnormalities implicating centromeric DNA". Nature Communications. 9 (1): 4340. Bibcode:2018NatCo...9.4340B. doi:10.1038/s41467-018-06545-y. PMC 6194107. PMID 30337534.
- ^ Neumann, Pavel; Navrátilová, Alice; Schroeder-Reiter, Elizabeth; Koblížková, Andrea; Steinbauerová, Veronika; Chocholová, Eva; Novák, Petr; Wanner, Gerhard; Macas, Jiří (2012). "Stretching the Rules: Monocentric Chromosomes with Multiple Centromere Domains". PLOS Genetics. 8 (6): e1002777. doi:10.1371/journal.pgen.1002777. PMC 3380829. PMID 22737088.
- ^ Dernburg, A. F. (2001). "Here, There, and Everywhere: Kinetochore Function on Holocentric Chromosomes". The Journal of Cell Biology. 153 (6): F33–8. doi:10.1083/jcb.153.6.F33. PMC 2192025. PMID 11402076.
- ^ Pluta, A. F.; MacKay, A. M.; Ainsztein, A. M.; Goldberg, I. G.; Earnshaw, W. C. (1995). "The Centromere: Hub of Chromosomal Activities". Science. 270 (5242): 1591–4. Bibcode:1995Sci...270.1591P. doi:10.1126/science.270.5242.1591. PMID 7502067.
- ^ a b Mehta, G. D.; Agarwal, M.; Ghosh, S. K. (2010). "Centromere Identity: a challenge to be faced". Mol. Genet. Genomics. 284 (2): 75–94. doi:10.1007/s00438-010-0553-4. PMID 20585957.
- ^ Dalal, Yamini (2009). "Epigenetic specification of centromeres". Biochemistry and Cell Biology. 87 (1): 273–82. doi:10.1139/O08-135. PMID 19234541.
- ^ Bernad, Rafael; Sánchez, Patricia; Losada, Ana (2009). "Epigenetic specification of centromeres by CENP-A". Experimental Cell Research. 315 (19): 3233–41. doi:10.1016/j.yexcr.2009.07.023. PMID 19660450.
- ^ Chueh, A. C.; Wong, LH; Wong, N; Choo, KH (2004). "Variable and hierarchical size distribution of L1-retroelement-enriched CENP-A clusters within a functional human neocentromere". Human Molecular Genetics. 14 (1): 85–93. doi:10.1093/hmg/ddi008. PMID 15537667.
- ^ Rosenfeld, Jeffrey A; Wang, Zhibin; Schones, Dustin E; Zhao, Keji; Desalle, Rob; Zhang, Michael Q (2009). "Determination of enriched histone modifications in non-genic portions of the human genome". BMC Genomics. 10: 143. doi:10.1186/1471-2164-10-143. PMC 2667539. PMID 19335899.
- ^ Chang, C-H; Chavan, A; Palladino, J; Wei, X; Martins, NMC; Santinello, B; et al. (2019). "Islands of retroelements are major components of Drosophila centromeres". PLOS Biol. 17 (5): e3000241. doi:10.1371/journal.pbio.3000241. PMC 6516634. PMID 31086362.
- ^ Volpe, T. A.; Kidner, C; Hall, IM; Teng, G; Grewal, SI; Martienssen, RA (2002). "Regulation of Heterochromatic Silencing and Histone H3 Lysine-9 Methylation by RNAi". Science. 297 (5588): 1833–7. Bibcode:2002Sci...297.1833V. doi:10.1126/science.1074973. PMID 12193640. S2CID 2613813.
- ^ Marshall, Owen J.; Chueh, Anderly C.; Wong, Lee H.; Choo, K.H. Andy (2008). "Neocentromeres: New Insights into Centromere Structure, Disease Development, and Karyotype Evolution". The American Journal of Human Genetics. 82 (2): 261–82. doi:10.1016/j.ajhg.2007.11.009. PMC 2427194. PMID 18252209.
- ^ Warburton, Peter E. (2004). "Chromosomal dynamics of human neocentromere formation". Chromosome Research. 12 (6): 617–26. doi:10.1023/B:CHRO.0000036585.44138.4b. PMID 15289667.
- ^ Thangavelu, P.U.; Lin, C.Y.; Vaidyanathan, S.; Nguyen, T.H.M.; Dray, E.; Duijf, P.H.G. (2017), "Overexpression of the E2F target gene CENPI promotes chromosome instability and predicts poor prognosis in estrogen receptor-positive breast cancer", Oncotarget, 8 (37): 62167–62182, doi:10.18632/oncotarget.19131, PMC 5617495, PMID 28977935
- ^ Zhang, W.; Mao, J-H.; Zhu, W.; Jain, A.K.; Liu, L.; Brown, J.B.; Karpen, G.H. (2016). "Centromere and kinetochore gene misexpression predicts cancer patient survival and response to radiotherapy and chemotherapy". Nature Communications. 7: 12619. Bibcode:2016NatCo...712619Z. doi:10.1038/ncomms12619. PMC 5013662. PMID 27577169.
- ^ Giunta, S; Funabiki, H (21 February 2017). "Integrity of the human centromere DNA repeats is protected by CENP-A, CENP-C, and CENP-T". Proceedings of the National Academy of Sciences of the United States of America. 114 (8): 1928–1933. doi:10.1073/pnas.1615133114. PMC 5338446. PMID 28167779.
- ^ "Centromere". Merriam-Webster Dictionary.
- ^ "Centromere". Dictionary.com Unabridged. Random House.
Further reading[edit]
- Mehta, G. D.; Agarwal, M.; Ghosh, S. K. (2010). "Centromere Identity: a challenge to be faced". Mol. Genet. Genomics. 284 (2): 75–94. doi:10.1007/s00438-010-0553-4. PMID 20585957.
- Lodish, Harvey; Berk, Arnold; Kaiser, Chris A.; Krieger, Monty; Scott, Matthew P.; Bretscher, Anthony; Ploegh, Hiddle; Matsudaira, Paul (2008). Molecular Cell Biology (6th ed.). New York: W.H. Freeman. ISBN 978-0-7167-7601-7.
- Nagaki, Kiyotaka; Cheng, Zhukuan; Ouyang, Shu; Talbert, Paul B; Kim, Mary; Jones, Kristine M; Henikoff, Steven; Buell, C Robin; Jiang, Jiming (2004). "Sequencing of a rice centromere uncovers active genes". Nature Genetics. 36 (2): 138–45. doi:10.1038/ng1289. PMID 14716315. Lay summary – Science Daily (January 13, 2004).
| Wikimedia Commons has media related to Centromere. |
No comments:
Post a Comment